fastq质量值转化
2010-04-15 13:44:15| 分类:
生物信息学
| 标签:
|举报
|字号大中小 订阅
http://blog.sina.com.cn/liuguiyou
http://blog.sina.com.cn/s/blog_4af3f0d20100gwra.html
fastq质量值有以下两种(实际上三种,PHRED,sanger和solexa,前两个相同):
第一种:sanger质量值
PHRED quality score of a base call, de?ned in terms of the estimated probability of error:
sanger质量值等于PHRED quality score:公式如下:
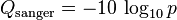
第二种:solexa质量值
The Solexa pipeline (i.e., the software delivered with the Illumina Genome Analyzer) earlier used a different mapping, encoding the odds ratio p/(1-p) instead of the probability p:

取值范围:
- Sanger format can encode a Phred quality score from 0 to 93 using ASCII 33 to 126 (although in raw read data the Phred quality score rarely exceeds 60, higher scores are possible in assemblies or read maps).
- Illumina 1.3+ format can encode a Phred quality score from 0 to 62 using ASCII 64 to 126 (although in raw read data Phred scores from 0 to 40 only are expected).
- Solexa/Illumina 1.0 format can encode a Solexa/Illumina quality score from -5 to 62 using ASCII 59 to 126 (although in raw read data Solexa scores from -5 to 40 only are expected)
换算关系:
- If the Phred quality is $Q, which is a non-negative integer, the corresponding quality character can be calculated with the following Perl code:
$q = chr(($Q<=93? $Q : 93) + 33);
where chr() is the Perl function to convert an integer to a character based on the ASCII table. - Conversely, given a character $q, the corresponding Phred quality can be calculated with: where ord() gives the ASCII code of a character.
- 同样的方法也可以应用于solexa数据
判断Sanger quality encoding或者solexa quality encoding:
The quickest way to distinguish Sanger Q-score encoding (ASCII-33) from Illumina (Solexa) Q-score encoding (ASCII-64) is to look for numerals [0-9] in the quality string. The numerals have ASCII values from 48-57 so it would be non-sensical to subtract 64 from them. If there are numerals in your quality string then the Q-score encoding is Sanger.
solexa质量值到sanger质量值的转化:
Conversion from ‘fastq-illumina’ to ‘fastq-sanger’ will be a common operation, and is very straightforward since both variants use PHRED scores but with di?erent o?sets. All that is required is to decrease the quality character codes by 31
参考资料:
1:http://en.wikipedia.org/wiki/FASTQ_format
2:Cock et al (2009) The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Research, doi:10.1093/nar/gkp1137http://nar.oxfordjournals.org/cgi/content/abstract/gkp1137 3:http://maq.sourceforge.net/fastq.shtml#intro
评论这张
 转发至微博
转发至微博
 转发至微博
转发至微博
关闭
玩LOFTER,免费冲印20张照片,人人有奖!
我要抢>
<#--最新日志,群博日志-->
<#--推荐日志-->
<#--引用记录-->
<#--博主推荐-->
<#--随机阅读-->
<#--首页推荐-->
<#--历史上的今天-->
<#--被推荐日志-->
<#--上一篇,下一篇-->
<#-- 热度 -->
<#-- 网易新闻广告 -->
<#--右边模块结构-->
<#--评论模块结构-->
<#--引用模块结构-->
<#--博主发起的投票-->

评论